Steps to making data reusable range from the organizational (document the contents of your files) to the complex (ensuring the computing environment of your data can be re-created). The first is sometimes more of a matter of convenience: a data file may be usable, but a fellow researcher cannot spare the many hours it might take to sort out how to use poorly documented materials. In other cases, badly documented data can be more than an inconvenience. It can make the entire dataset unusable by you or your colleagues because the context for how the data was collected or how it models your real-world referent cannot be recalled.
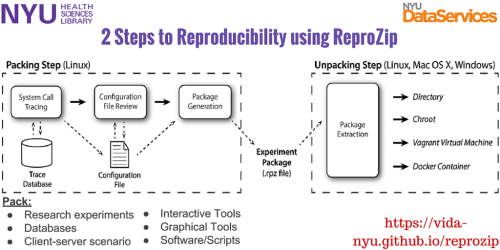
For the second case, researchers and developers are working hard to build options for capturing the computing environment in which data is built or analyzed. For help in finding those tools, take a look at resources like ReproMatch, a searchable list of reproducibility tools. Or give tools like ReproZip a try. ReproZip is a tool developed by the ViDA (Visualization and Data Analysis) group at NYU to make reproducibility easy. It tracks operating system calls and creates a package that contains all the binaries, files, and dependencies required to run a given command on the original researcher’s computational environment. A reviewer can then extract the experiment in his own environment to reproduce the results, even if the environment has a different operating system from the original one.
Another good place to start to follow developments in using and re-using data is Reproducible Science, a consortium arising out of the Moore/Sloan Data Science Environment and its partners, New York University, the University of California at Berkeley, and the University of Washington.
